geocomplexity for spatial raster data based on spatial dependence
Source:R/geocd_raster.R
geocd_raster.RdThis function calculates geocomplexity for spatial raster data based on spatial dependence.
Arguments
- r
SpatRasterobject or can be converted toSpatRasterbyterra::rast().- order
(optional) The order of the adjacency object. Default is
1.- normalize
(optional) Whether to further normalizes the calculated geocomplexity. Default is
TRUE.- method
(optional) In instances where the method is
moran, geocomplexity is determined using local moran measure method. Conversely, when the method isspvar, the spatial variance of attribute data serves to characterize geocomplexity. For all other methods, the shannon information entropy of attribute data is employed to represent geocomplexity. The selection of the method can be made from any one of the three options:moran,spvarorentropy. Default ismoran.
Note
In contrast to the geocd_vector() function, the geocd_raster() performs operations
internally on raster data based on neighborhood operations(focal) without providing
additional wt object.
References
Zehua Zhang, Yongze Song, Peng Luo & Peng Wu (2023) Geocomplexity explains spatial errors, International Journal of Geographical Information Science, 37:7, 1449-1469, DOI: 10.1080/13658816.2023.2203212
Anselin, L. (2019). A local indicator of multivariate spatial association: Extending geary’s c. Geographical Analysis, 51(2), 133–150. https://doi.org/10.1111/gean.12164
Examples
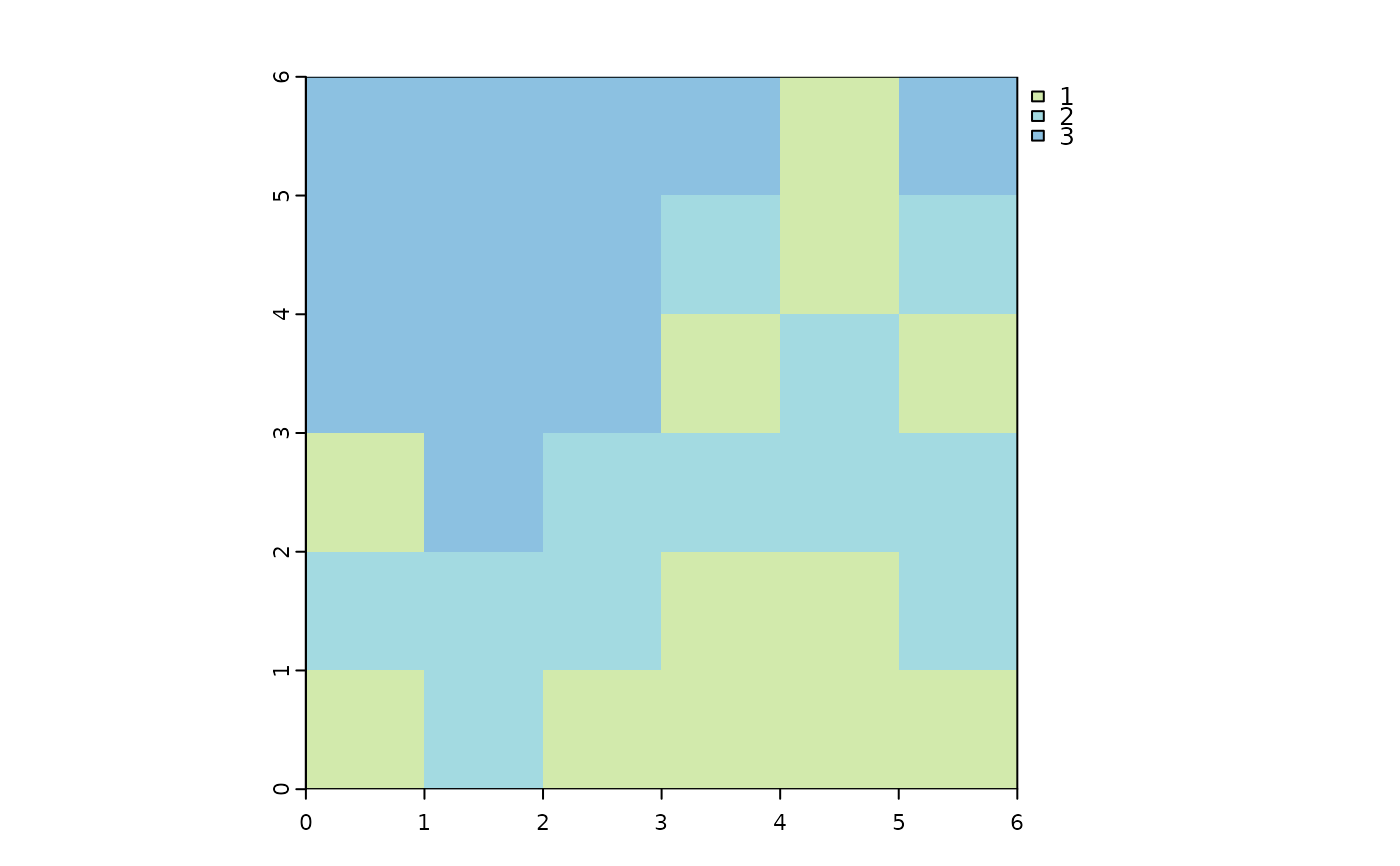
library(terra)
#> terra 1.8.60
m = matrix(c(3,3,3,3,1,3,
3,3,3,2,1,2,
3,3,3,1,2,1,
1,3,2,2,2,2,
2,2,2,1,1,2,
1,2,1,1,1,1),
nrow = 6,
byrow = TRUE)
m = rast(m)
names(m) = 'sim'
plot(m, col = c("#d2eaac", "#a3dae1", "#8cc1e1"))
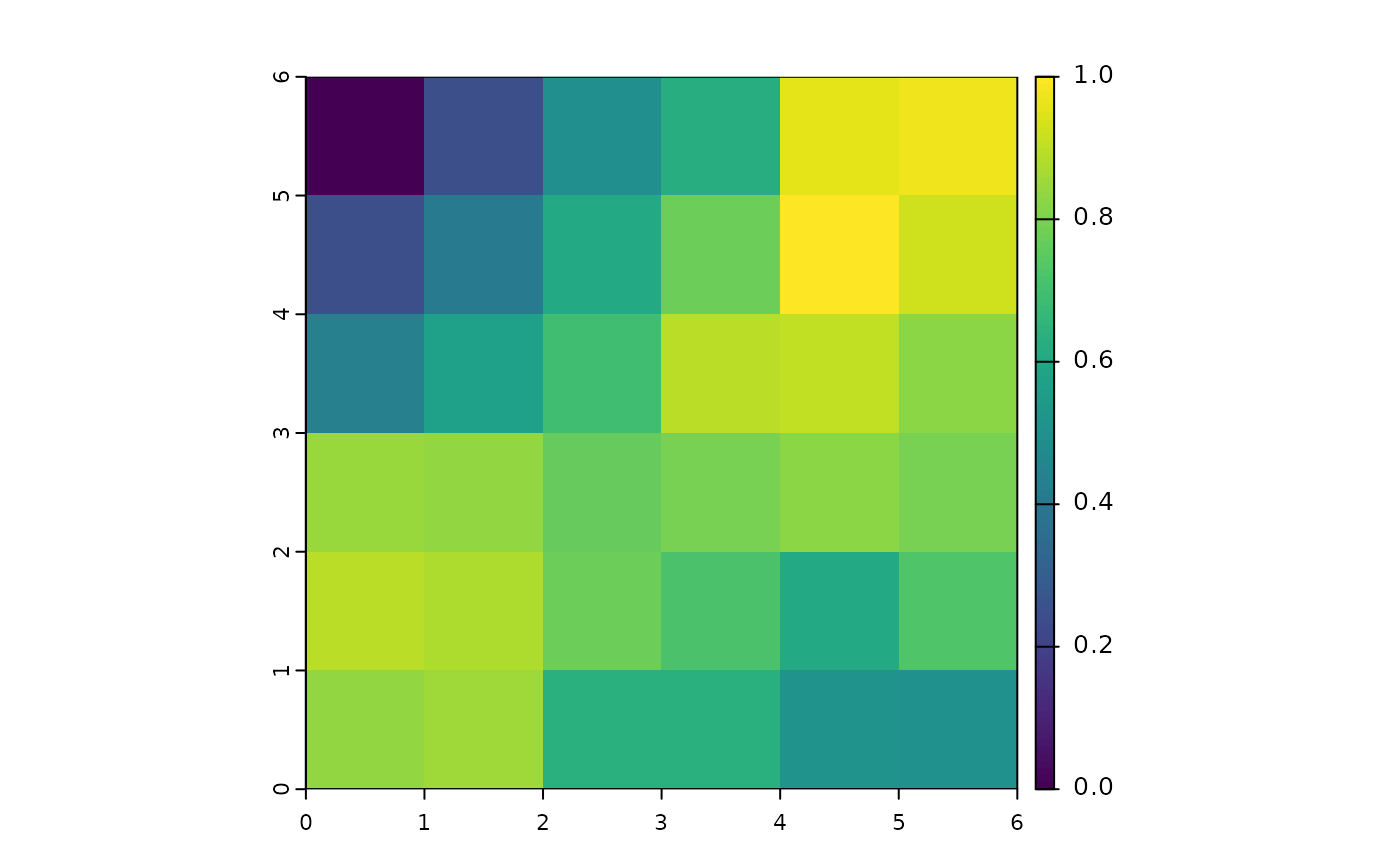
 gc1 = geocd_raster(m,1)
gc2 = geocd_raster(m,2)
gc1
#> class : SpatRaster
#> size : 6, 6, 1 (nrow, ncol, nlyr)
#> resolution : 1, 1 (x, y)
#> extent : 0, 6, 0, 6 (xmin, xmax, ymin, ymax)
#> coord. ref. :
#> source(s) : memory
#> name : GC_sim
#> min value : 0
#> max value : 1
plot(gc1)
gc1 = geocd_raster(m,1)
gc2 = geocd_raster(m,2)
gc1
#> class : SpatRaster
#> size : 6, 6, 1 (nrow, ncol, nlyr)
#> resolution : 1, 1 (x, y)
#> extent : 0, 6, 0, 6 (xmin, xmax, ymin, ymax)
#> coord. ref. :
#> source(s) : memory
#> name : GC_sim
#> min value : 0
#> max value : 1
plot(gc1)
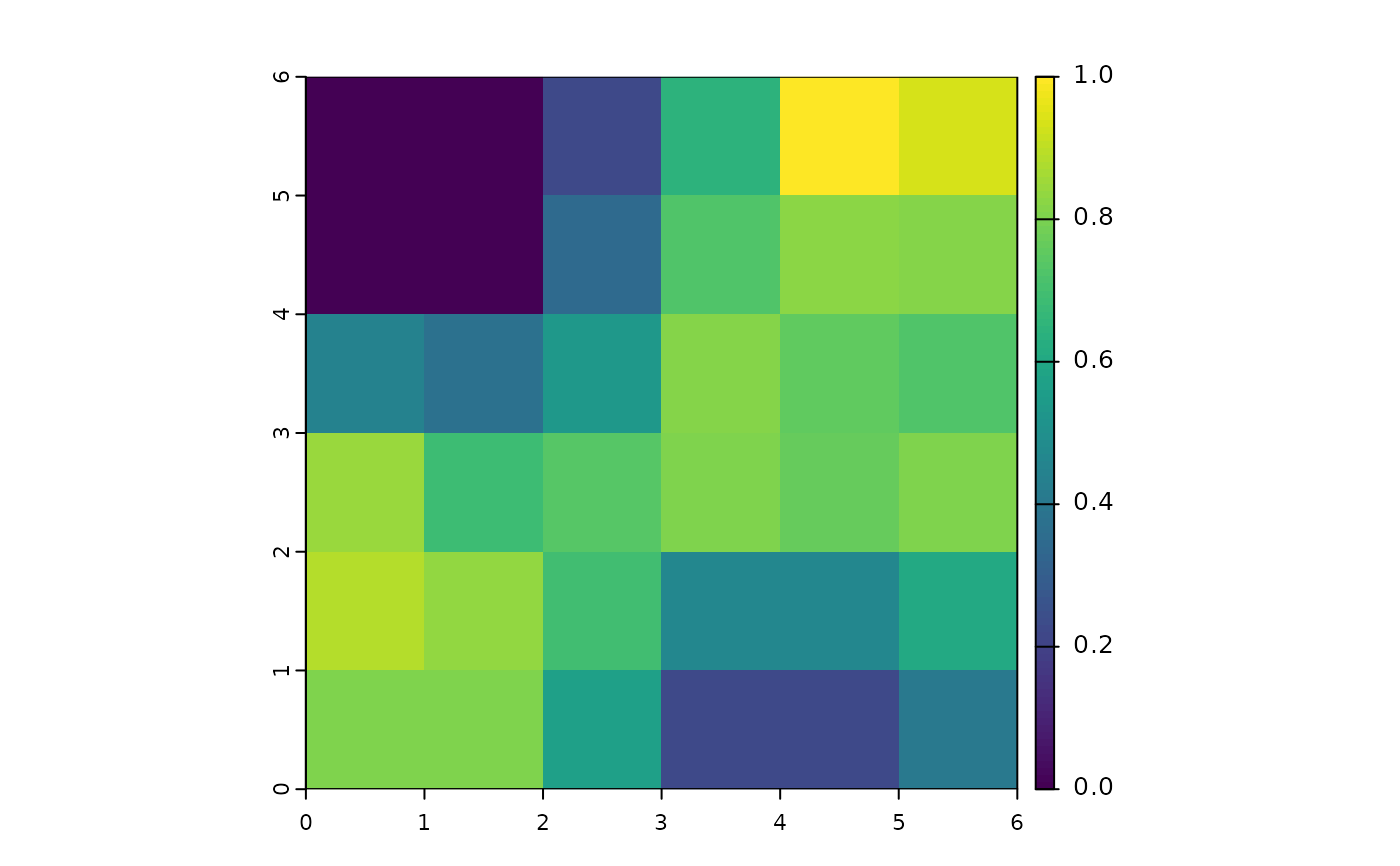 gc2
#> class : SpatRaster
#> size : 6, 6, 1 (nrow, ncol, nlyr)
#> resolution : 1, 1 (x, y)
#> extent : 0, 6, 0, 6 (xmin, xmax, ymin, ymax)
#> coord. ref. :
#> source(s) : memory
#> name : GC_sim
#> min value : 0
#> max value : 1
plot(gc2)
gc2
#> class : SpatRaster
#> size : 6, 6, 1 (nrow, ncol, nlyr)
#> resolution : 1, 1 (x, y)
#> extent : 0, 6, 0, 6 (xmin, xmax, ymin, ymax)
#> coord. ref. :
#> source(s) : memory
#> name : GC_sim
#> min value : 0
#> max value : 1
plot(gc2)